How to Create and Animate Chemical Structures in 3D using Online Databases – Part I
Last updated on 4 March 2024, Published on 13 February 2023
This 2-part series of articles offers different options to the content creators for making chemical structures in 3D for their use in audiovisual content. Here, we’re using Jmol (with step-by-step guidelines) and other programs to show how you can create chemical structures and then use them in programs like After Effects and 3ds Max for animations. Also, we’re diving deeper into different online chemical databases showing how you can obtain molecules along with data and structures.
Introduction
We often need to make chemical structures in 3D for various scientific and commercial presentations, especially through explainer videos.
Be it for an e-learning courseware or conventional educational program or for a pharmaceutical company’s explainer video on the utility of a component, using 3d chemical structures in videos is indispensable for the obvious purpose it serves.
Any explanation relating to chemistry demands some sort of molecular and atomic structures in the presentation for a better understanding of the viewers. Therefore, it’s important that the content creators, especially the video makers, find a solution for making chemical structures in 3d in a fast and convenient way.
It may look daunting for a content creator to present a 3d simulation of molecular structures right at the outset. But a methodical breakdown and step-by-step approach to the workflow will lead you to a presentation that’s elegant and professional.
Even for the game developers, especially the modelers, a knowledge of building chemical structures in 3d may be useful at some point in time.
Here are two things we have to tackle. One is the science of the very knowledge on a particular topic. Second, the very scene is set up following the topic content.
This two-part series of articles will help you on two different counts. The programs that help us create the molecules, build a scene and animate. And also, we will look into using free online resources for the content.
We shall be using three different programs Jmol, 3ds Max and Adobe After Effects for creating and animating the chemical structures in 3d.
We shall also be using Protein Data Bank (PDB) like RCSB PDB or PubChem database to use different molecular structures. It may be helpful for many of us who don’t usually have to tread the arena of hardcore science.
Here is the list of topics to be covered in this article i.e Part I.
- What is Jmol?
- How to create chemical structures in 3D using Jmol?
- What next? – dive deeper into Jmol and export
- A simple composite in After Effects with exported image [from Jmol]
- How to get 3D chemical structures from online databases like RCSB PDB & PubChem?
What is Jmol?
Jmol is an open-source and free program for creating three-dimensional chemical structures. Widely used by the students and teachers fraternity, the application runs on Windows, Mac, and Unix (Linux) systems. You can download this stand-alone program easily from Sourceforge.
For the purpose of this article, we shall be using Jmol 16.1.1. The above download link as of writing the article is showing version 14 as the current release, while it actually gets version 16.1.3.
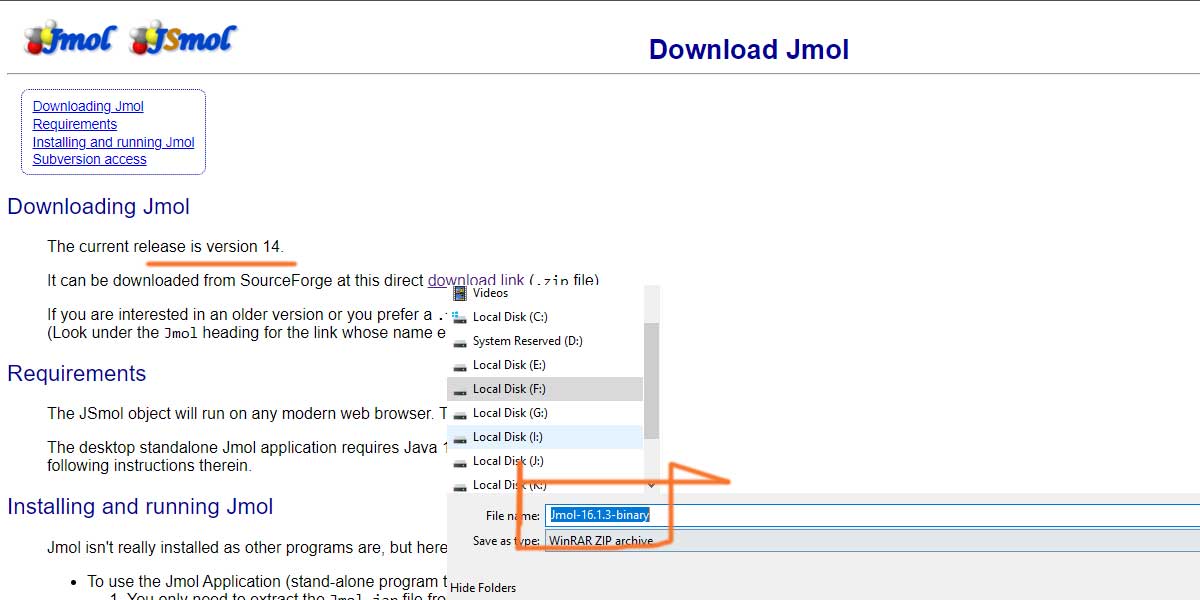
The installation preceded by the download is a simple process and it will finally take you to a folder with Jmol with an executable jar file within it. You only have to double-click on it. Isn’t that wonderful!
Before we go on further for taking a quick tour inside Jmol, it’s important to take a few additional notes regarding the program. There is something called JSmol that can be used along with Jmol. This application or JSmol is a web browser that’s used for online courseware. It’s integrated into webpages and calls for web-accessible chemical databases like Protein databank or PDB (more on this online chemical database later).
JSmol is a JavaScript framework that helps developers integrate chemical structures in 3d in webpages or devices without having to install Java. Once JSmol is integrated, Jmol can display interactive 3d chemical structures there without the need of installing Java.
There is a minor difference between JSmol with HTML 5 and JSmol with Java. Java makes JSmol render faster than the program with HTML 5 and without Java. But that’s hardly noticeable for the chemical structures with less than 20 thousand atoms.
For our purpose here, we shall be using Jmol i.e. the stand-alone application launch on double clickings, like tons of other applications.
How to create a 3d chemical structure in Jmol?
After launching Jmol, you would land on a somewhat ancient-looking interface, similar to other old applications written in Java. But that’s precisely what it makes simple and lightweight.
After launching the program, your work area may look different than mine. Two ways to make the display area bigger or fit your monitor. One is to maximize the work area by clicking on the top right corner.
Another way is to Go to Display on the Menu > Click on Resize and put the values you want.
Since we’re not dedicated to learning Jmol in its entirety here, we shall instead focus on creating molecules either from scratch or importing from online databases fast, essentially jumping over at times without touching every nook and corner.
For online courseware on the basic chemistry lessons, you may have to start with the Model Kit as shown. It will immediately pop up methane CH4. This Model Kit helps create molecules with ‘minimized energy’ i.e. in the most stable configuration. And that’s important because a faculty member can ask you to create a compound in its most stable configuration.
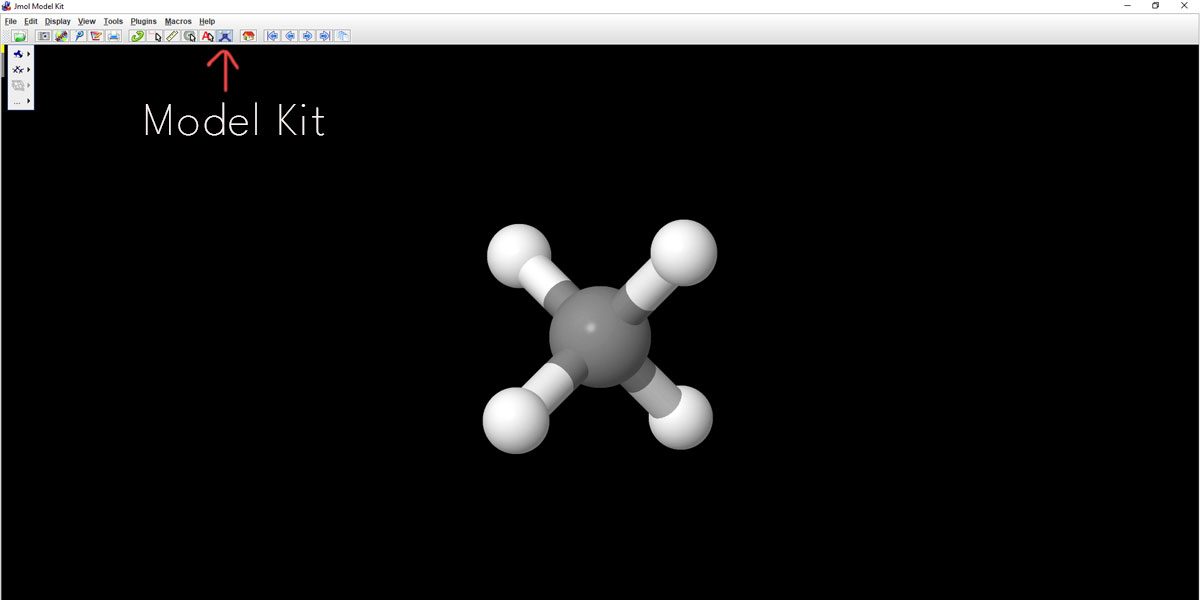
With a few more steps, you could be able to add or delete some functional groups to this or any compound. Click on the Model Kit and you’ll get to add many different atoms or functional groups as per requirement. Take a pause and get a quick tour of the different features offered here in the Model Kit.
If you take your cursor to one of the Hydrogen atoms in Methane, it will turn red and once you click, the Hydrogen atom will be replaced by one methyl group i.e CH3. Here we can see Methane is changed into Ethane.
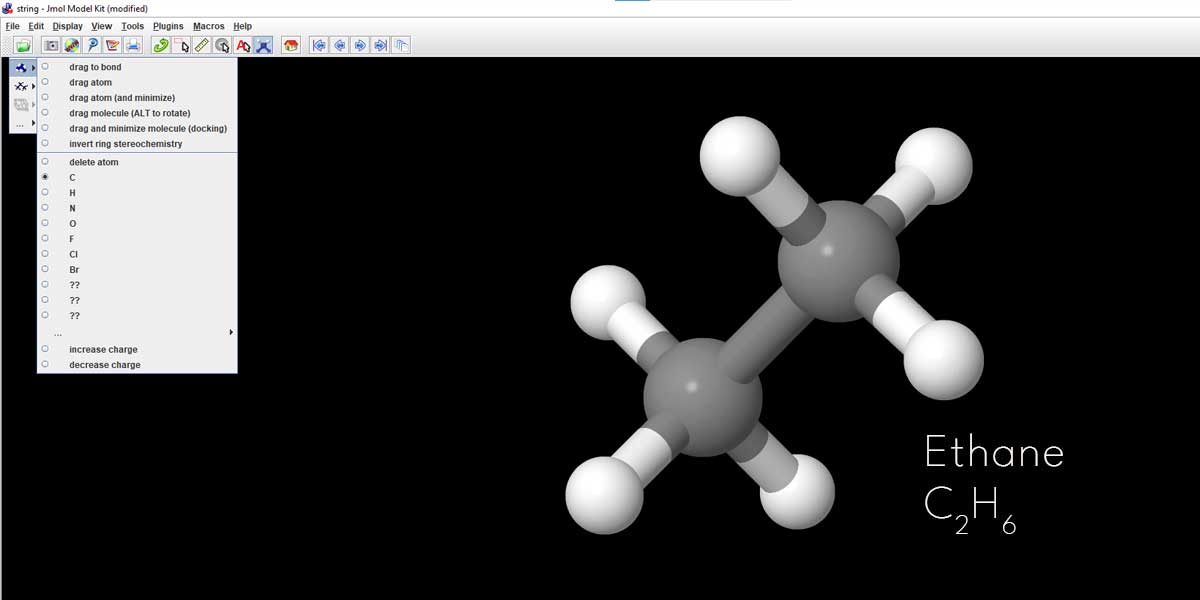
We just had a talk about energy minimization for attaining the most stable configuration of the molecule. Here is an example of turning a Propane molecule to its most stable state. Again, this Propane we created by clicking on one Hydrogen atom of the Ethane in the previous example.
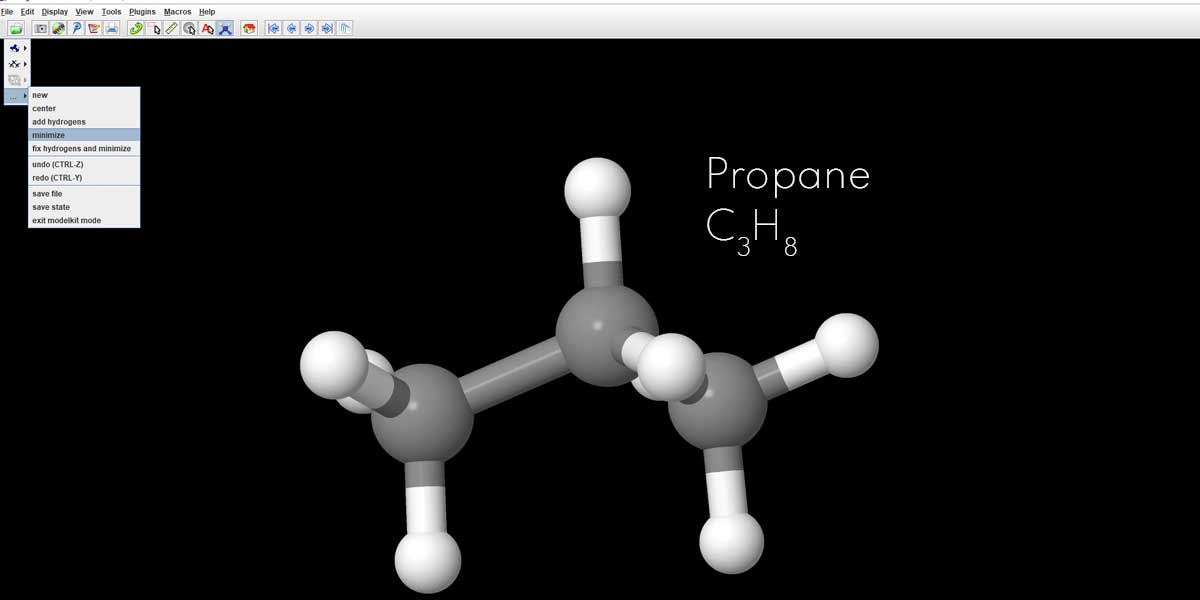
In the Model Kit, scroll down to the bottom and just click on minimize. You’ll see the molecule instantly adjusts its configuration with the sub-atomic distances re-adjusted taking it to the most stable state.
There are many ways you can tweak this model kit and other tools to edit the molecular structures before exporting it to a format compatible with a 3d program. You can add and delete functional groups from the Model Kit of Jmol while making your chemical structures in 3d.
Using a simple trick, you can convert this Propane model to a Propanol molecule.
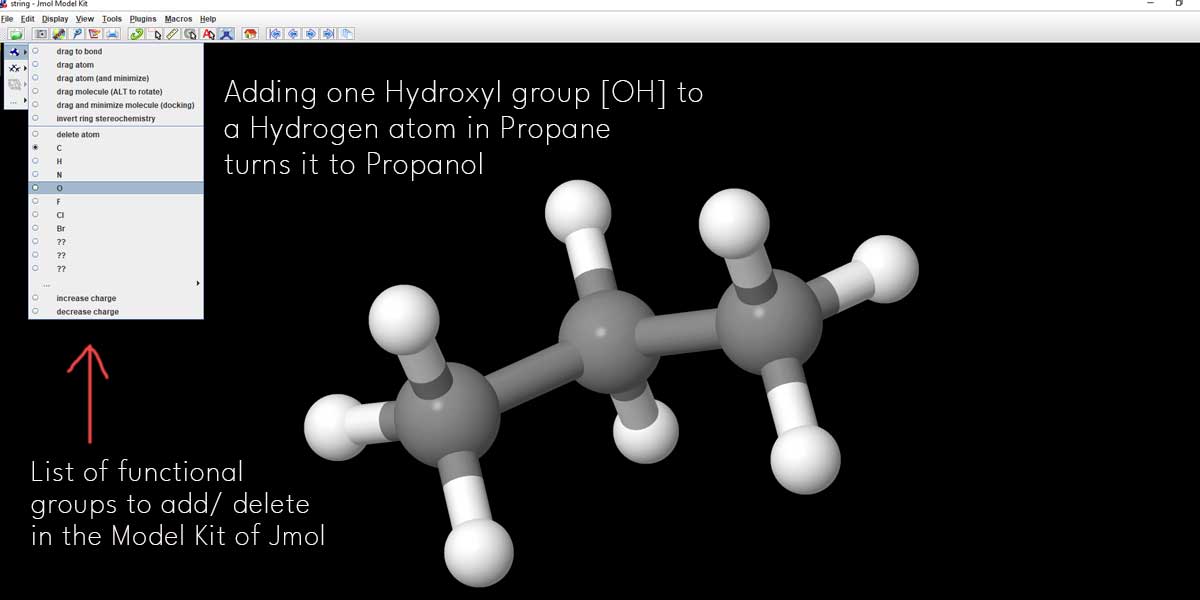
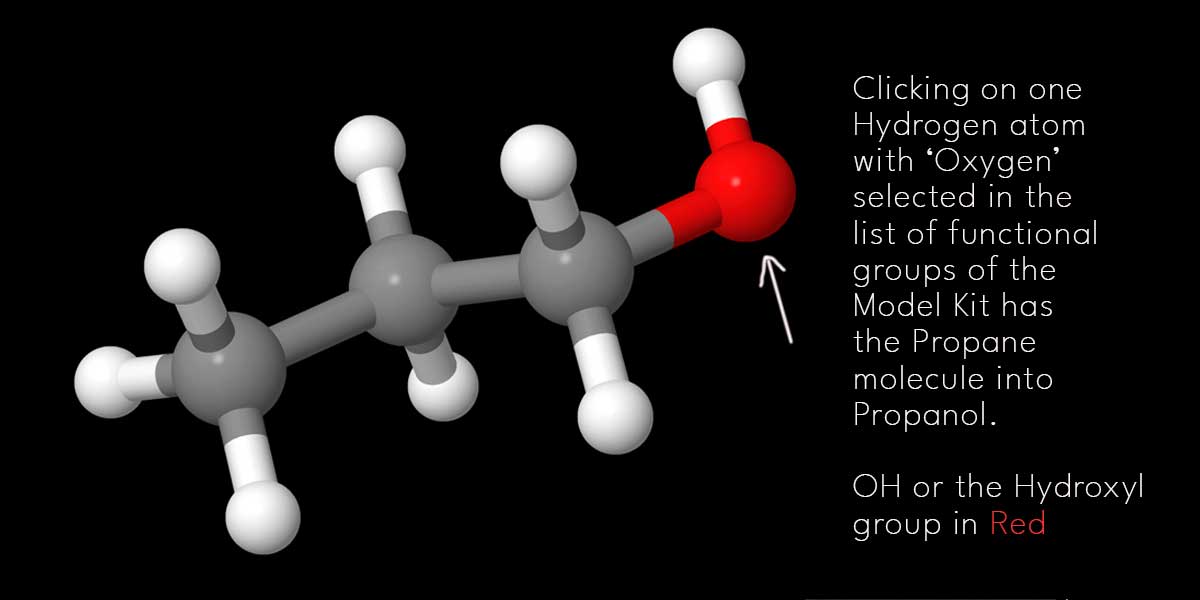
If you add one Hydroxyl group replacing one Hydrogen atom, you can convert Propane to Propanol. There you can see a list of functional groups available there. Click on the check box left to O or Oxygen. Then go to the first Carbon atom [extreme right or left] and click on the Hydrogen atom associated with the ‘first Carbon atom’.
The Hydrogen atom will be immediately replaced by one Hydroxyl group or OH. You can see the new Hydroxyl group in red.
Before we further proceed, it’s to mention here that this is no chemistry lesson or Jmol tutorial guide either. Once you, as a content creator, get to the idea of how to create basic chemical structures using Jmol and its Model Kit, it’s high time you leave this program to animate the molecules in a different program.
We won’t animate the chemical structures here.
What next – dive deeper into Jmol & export?
So far you get to how you can create basic chemical structures in 3d and edit their functional groups. You can also take a deeper dive by exploring the program features. There are plenty, in fact. You can measure subatomic distance, change colors or different atoms, bonds, etc.
But before exporting any molecule, let’s say, the Propanol or propane we just made from Jmol, we would like to have a look at two features that will help you get molecules from online sources. Jmol can access chemical databases like PubChem or RCSB Protein Data Bank to obtain different molecular structures.
Now, these chemical structures are certainly not at the basic level meant for eighth-grade chemistry students. There are millions of compounds listed in PubChem or PDB that one can hardly create from scratch. That’s why you may have to get the entries from these databases and create the chemical structures in 3d.
Jmol has an option whereby you can access these databases. Let’s now see how you can do that.
Go to File > Click on Get Mol
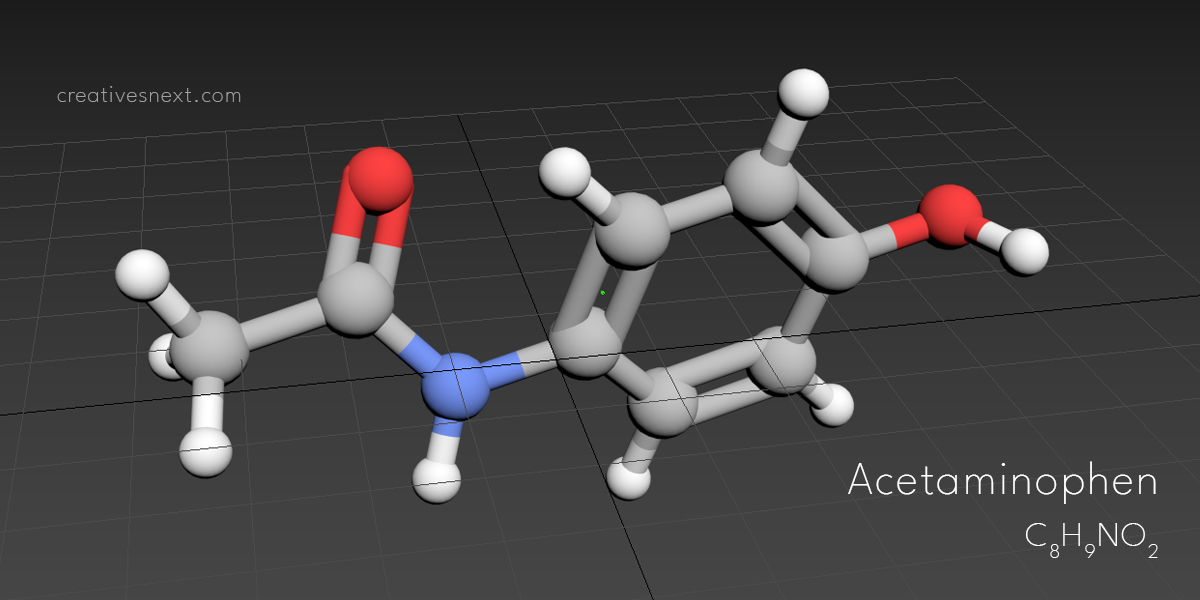
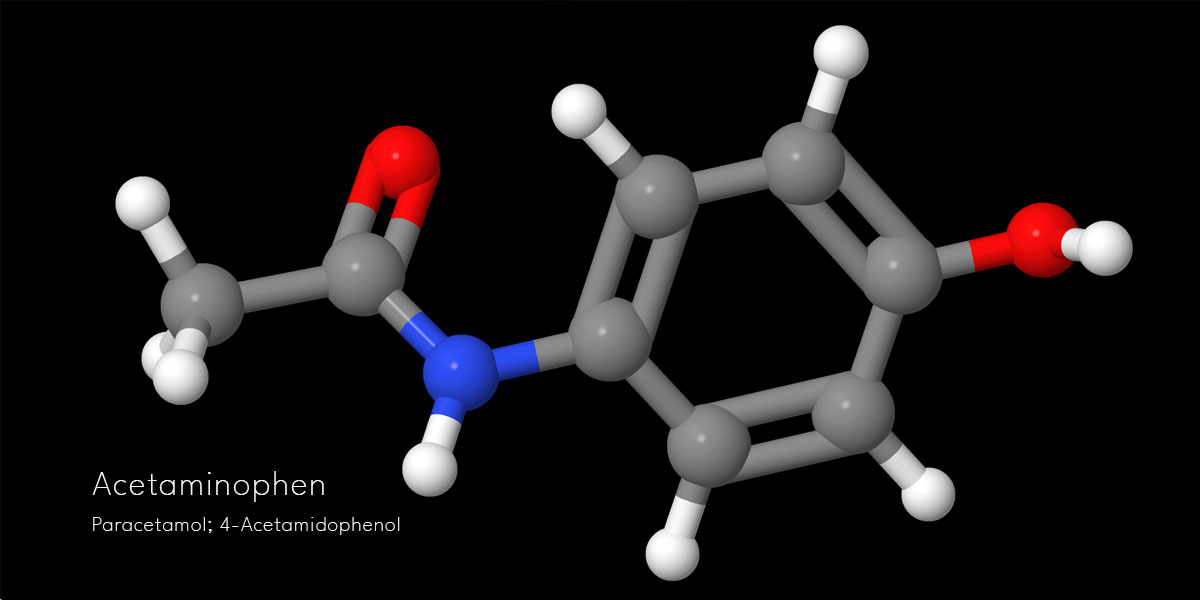
Let’s now put a relatively bigger molecule that you saw in this post’s featured image – Acetaminophen. It’s a commonly used compound used in Pharma Industry for making Paracetamol [4-Acetamidophenol; HOC6H4NHCOCH3]. Just type in Acetaminophen in the input dialogue box that popped up after clicking on the ‘Get Mol’.
Here the idea is to get such complex chemical structures in 3d from a database and create that inside Jmol. Once that’s done you are left with just one step to get out of Jmol i.e. export.
Let’s see what comes up with our Acetaminophen!
You can also use Get PDB featured under the File menu to access the Protein data bank. We shall see one instance before we prepare our chemical structures in 3d for export.
This time Scroll down to Get PDB and you’ll see an input dialogue box appears immediately. Here the task is a bit different. You have to put a four-letter Protein Id from the Protein Data bank.
The Protein Databank is a widely used International database for Protein and nucleic acid molecules. All you have to do is visit the site, find the protein molecule of your interest, and then find its respective four-letter id.
Let’s type Human Enzyme in the search field of the protein databank website and see what happens. We shall come back later on these two repositories viz. PubChem and RCSB PDB to have a wider look. But for now, let’s limit ourselves to how to get the protein id from this PDB and put that four-letter id in the input dialogue box that pops up after clicking on the Get PDB sub-menu.
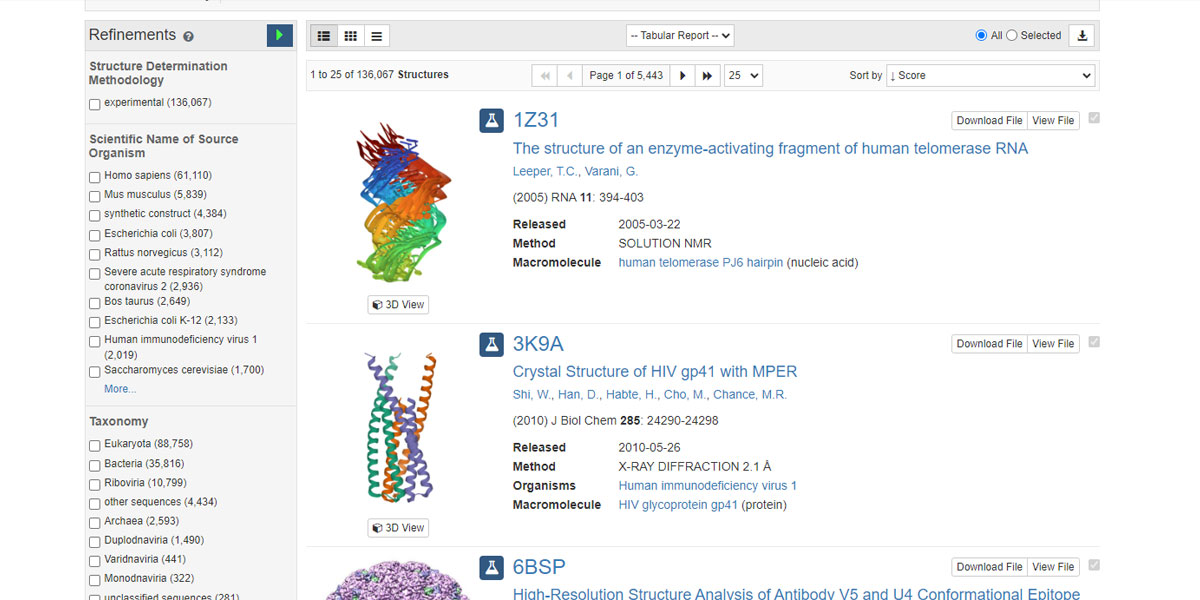
You see there are four-letter ids against each entry. Now you have to pick one such id and paste that into the input dialogue box. Let’s put 1Z31 in the input dialogue box and see what happens.
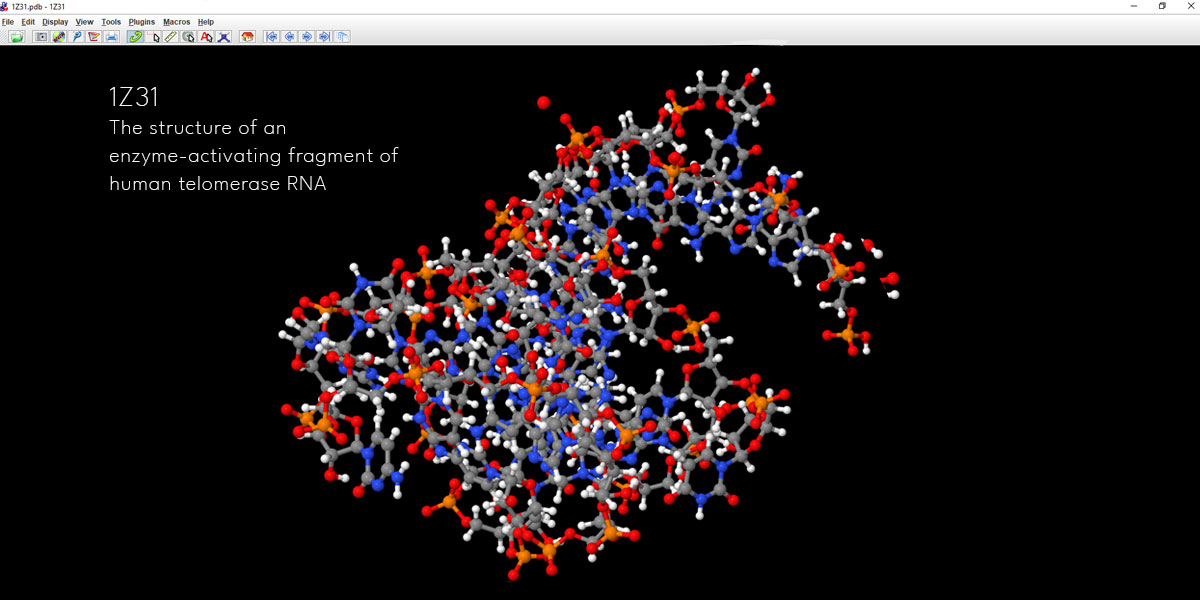
This is certainly not something to create from the scratch. So if you’re into creating a Pharma company’s presentation on their certain drug molecule whereby you’re to create the 3d chemical structures something like this, you can surely now visit these two databases and find a molecule of your client’s interest.
We’ll visit these two sites later to find out what they offer and how you can make the best use of them. But for now, you will look into how to get the chemical structures in 3d from Jmol for their use in a 3d program like 3ds Max. Essentially, how to export them.
Let’s get back to our Propanol or Propane file. It’s important to exit the Model Kit mode. Unless you exit the Model Kit you won’t able to export in the way you like it to. So exit from the Model Kit as shown in the image below.
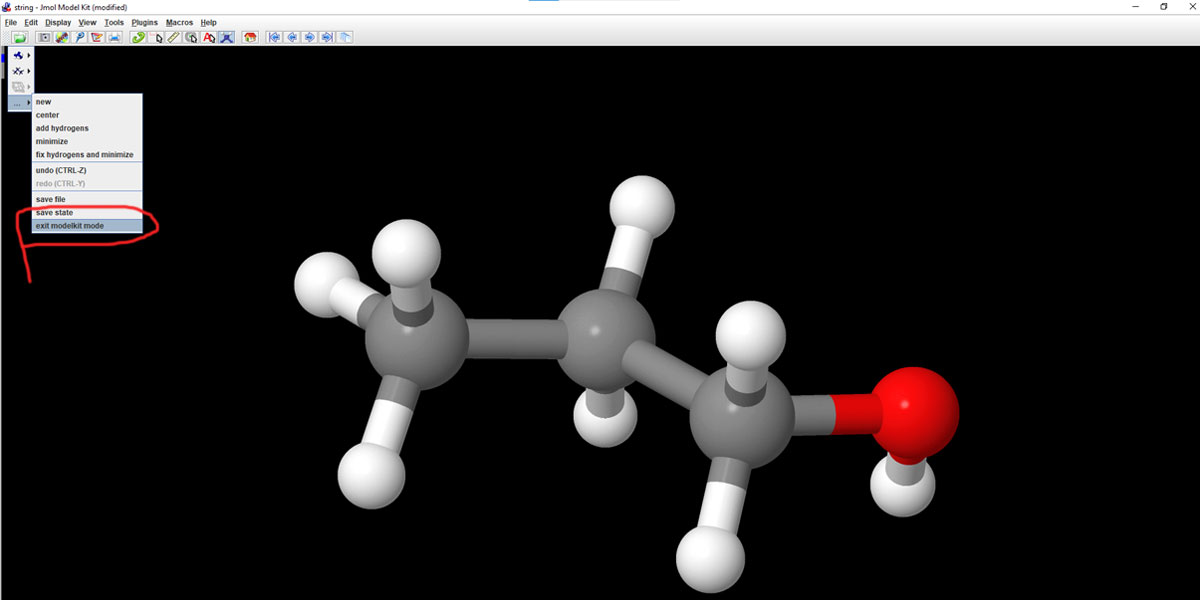
Even while within the Model Kit mode, you can access the Export option from the File menu, but that doesn’t offer certain features necessary for 3d program compatibility.
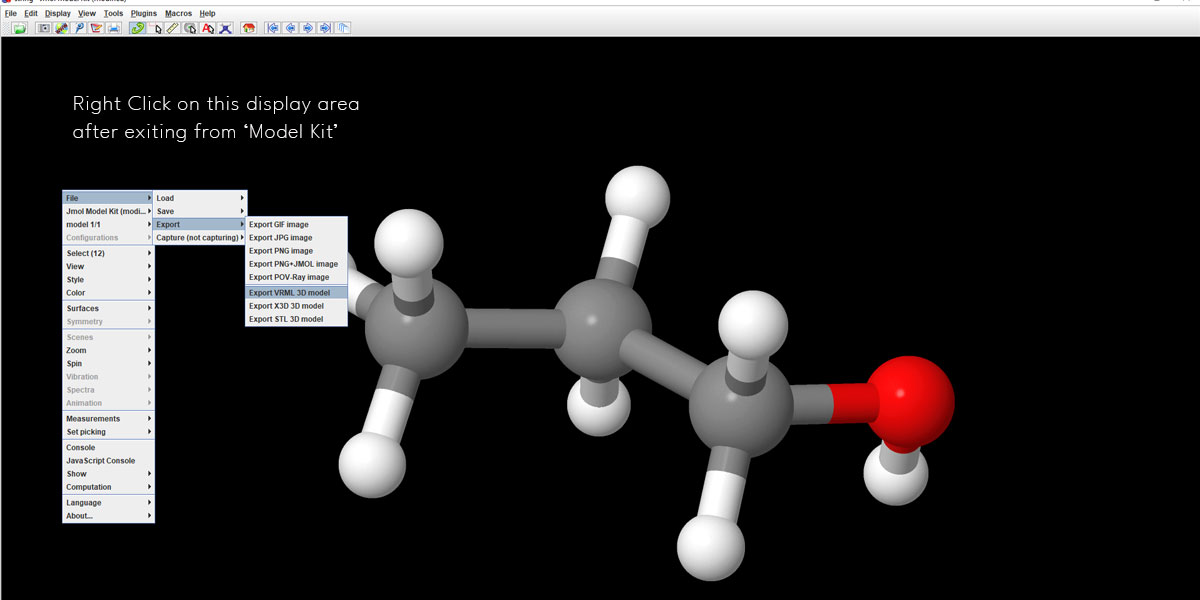
After exiting the Model Kit mode, right-click anywhere in the display area (except the structure) and you’ll get to see the Export option under File. Select VRML 3D model to export your structure.
VRML is the Virtual Reality Modeling Language. Jmol offers most of its features for a 3d structure via this format. You can export the file in STL or x3D too. But VRML is cross-platform and is compatible with the 3D printing industry.
We can export a simple image in PNG or JPG from Jmol too and work on that to create simple animations that can be used in basic chemistry lessons. This could be helpful for the YouTubers who create tons of videos for schoolgoers. So before diving into 3ds Max, let’s do a simple composite with Adobe After Effects with a PNG image
A simple composite in After Effects with exported image [from Jmol]
Again, right-click on any point except the structure and select File > Export > Export PNG Image. We save our files in PNG. It doesn’t provide transparency with Alpha though. But that’s no issue. Let’s head over to After Effects. We’ll fix it there.
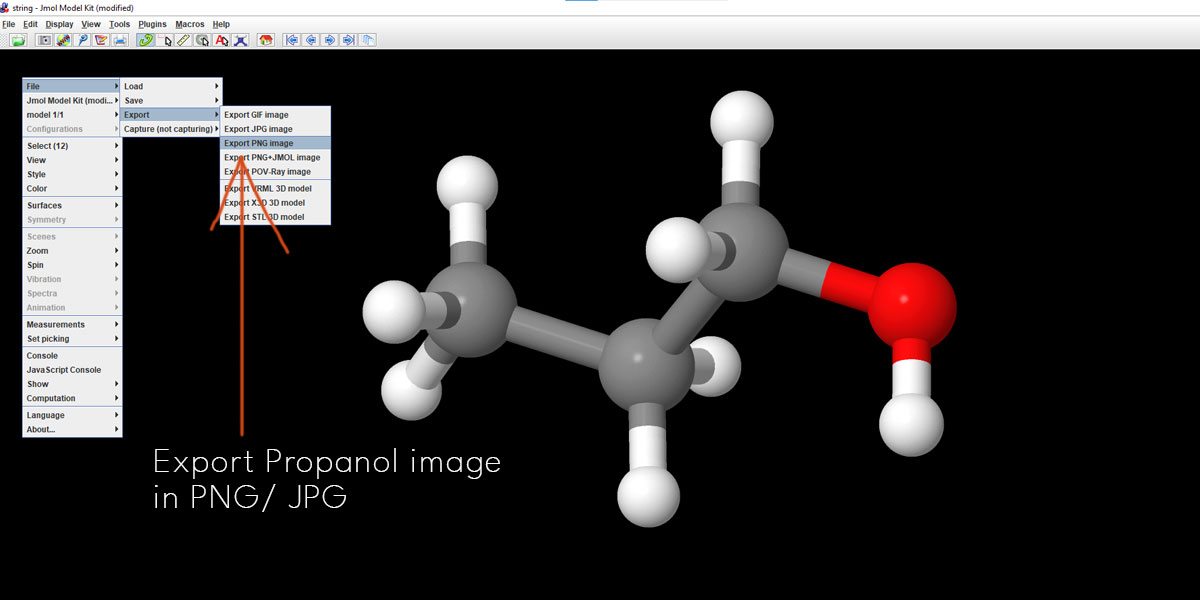
Let’s first see what we have come up with on After Effects. Here is a 10-second clip of what we could do with a few simple steps. Below are the breakdowns.
- Import PNG into an AE Composition. We had a 10-sec comp of 1920X1080 with FPS @ 29.97
- Jmol doesn’t give you Alpha, so use Linear Color Key to key out black in the image.
- Create a simple Grid, preferably with a dark-colored background in contrast with the molecular structure.
- Finally, turn the keyed-out layer of PNG into 3D and add wiggle expression to Position X, Y, Z, and rotation. You may try out a few combinations.
Just in case, you’re interested to have a sneak peek of ours, here is a reference.
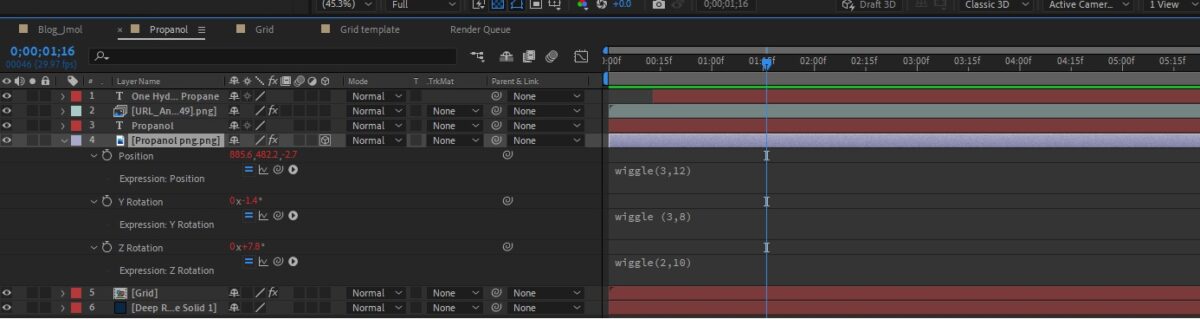
Now, this simple wiggle animation with the PNG image on After Effects doesn’t require you to dive into a 3d program like Blender or 3ds Max. Although After Effects is no 3D program and 2.5D only, basic-level chemistry courseware could use this sort of animation. This animation may look simple, but it helps the content creators while working on the explainer videos along with a host or speaker explaining chemistry lessons to the kids.
The problem of only using After Effects without any 3D program is you can’t rotate the molecule in Z space. For that, we have to use a 3D program.
But, before using the 3D program, let’s have a wider look at the online chemical databases, like PDB or PubChem that continue to help users all over the World with their huge pool of resources.
How to get 3D chemical structures from online databases like RCSB PDB & PubChem?
There are many online databases available for chemical structures that offer large biological molecules such as proteins, nucleic acids, and other complex organic compounds. Two of the most prominent and widely used websites are rcsb.org and PubChem. All over the World, people in the domain of biomedical science, Pharma, and Biotechnology extensively use these two websites to collect data and structures.
PDB (popularly known as) stands for Protein Data Bank. The website rcsb.org provides an international open-access database for protein biological molecules and nucleic acids. This database offers three-dimensional structural data of large biological molecules. It’s pivotal for the people in research.
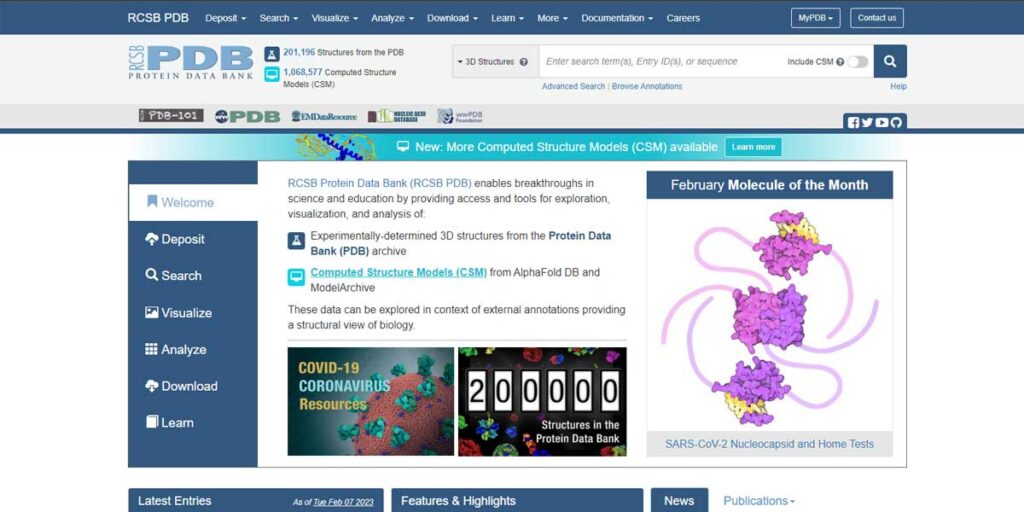
education and other commercial domains where the use of chemical structures in 3D is often required.
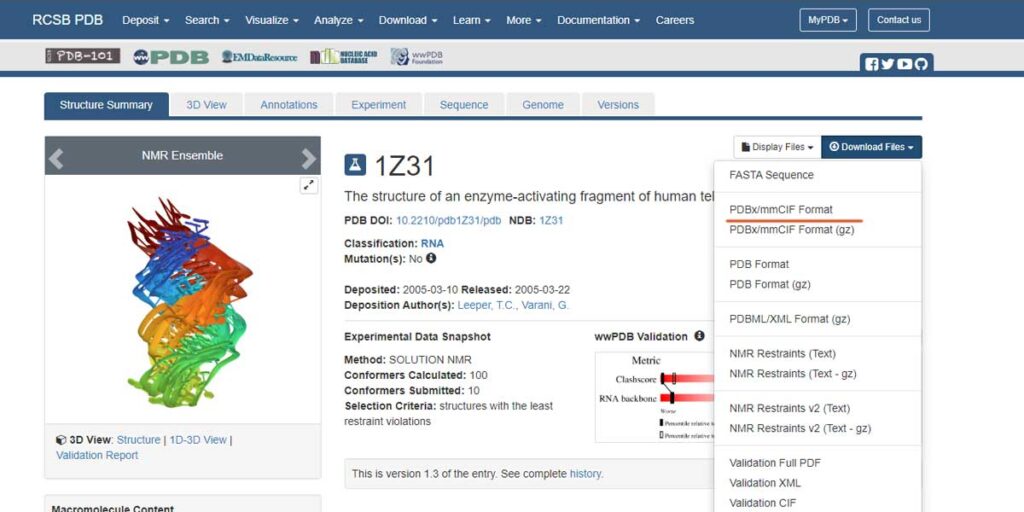
For any structure, you have to get the respective PDB id. The site is updated weekly. The file format of the molecules was originally PDB, which was later standardized as mmCIF. This format is acceptable to different open-source programs like Jmol, Pymol, Rasmol and etc. This article shows how you can access Protein Data Bank and get your molecules using the Get PDB sub-menu.
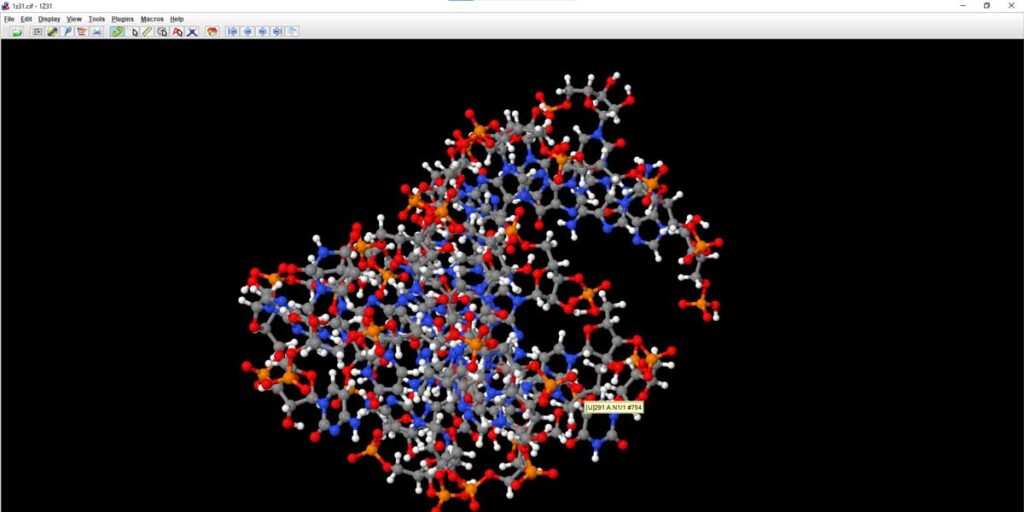
Alternatively, you can access the database directly by typing the 4-letter PDB id, in this case again 1Z31. Under Download Files, you’ll get many acceptable formats including mmCIF and PDB formats. Let’s download the mmCIF format and head over to Jmol to open it.
mmCIF is an acceptable format in Jmol, so no issue opening it from Open dialogue, just in case you have the file and you’re not connected to the internet. And, within a second you land up on the structure of an enzyme-activating fragment of human telomerase RNA [PDB Id 1Z31].
PubChem is a US Govt website, which is maintained by the National Center of Biotechnology Information, a component of the National Library of Medicine. This site has millions of organic compound structures and data that can be freely accessed and downloaded.
PubChem provides both image and 3D-compatible formats for downloading. Unlike, PDF you have to know a popular chemical name, like Toluene or Aspirin or IUPAC name to find the molecule of your choice.
Here, we have chosen Aspirin. You can see there is a downloadable format called SDF out of quite a few. We can download this .sdf file or click on the Get Mol submenu of Jmol.
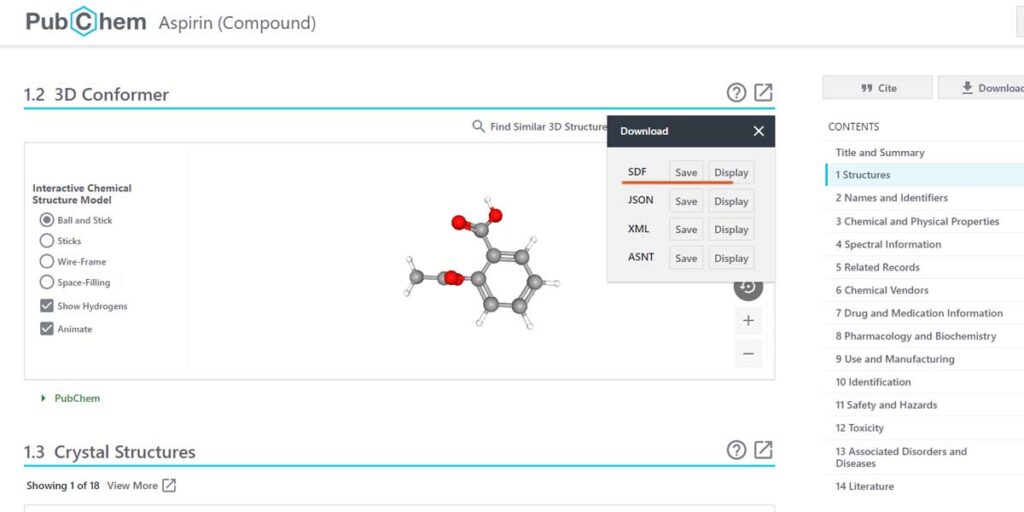
We download the.SDF file and then open it within Jmol. Either way, you’ll land up on the same Aspirin structure. Cool.
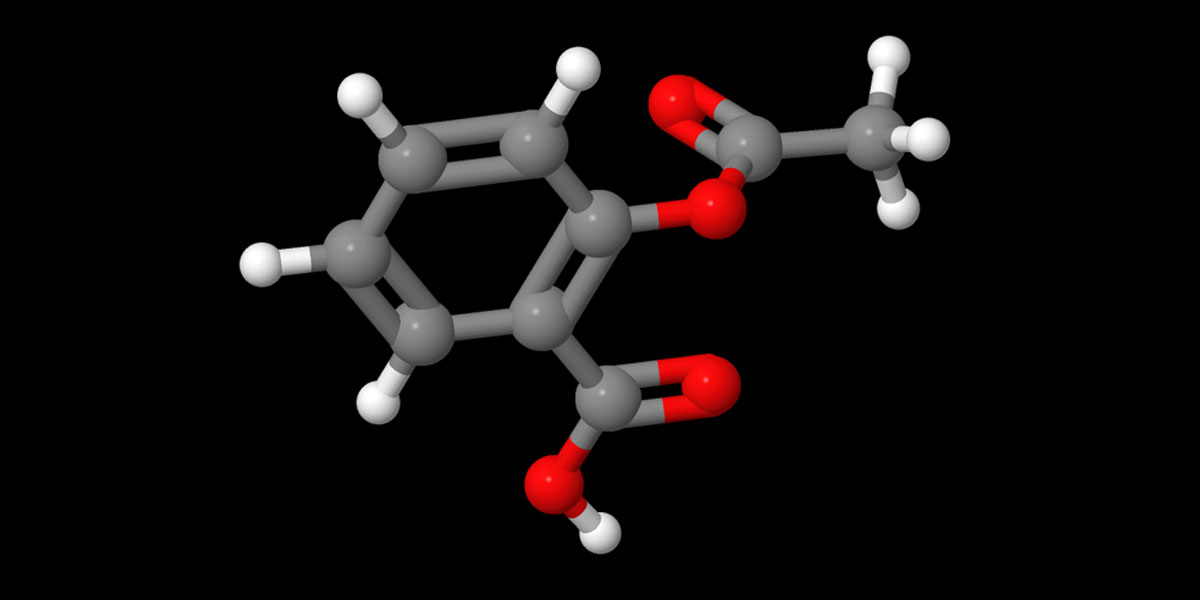
So you know how to use two huge databases for any chemical structures in 3D that you can use in our audio-visual content.
Please share the article and help me write more such pieces of content.
